COSC241: Lectures 22 and 23
Biological Data Science
Alex Gavryushkin
![]()
17 and 21 May 2018
Handouts
- All slides are online at
- You can follow this lecture at
Outline
1. Data Science (my take)
ML = Machine Learning
CoSt = Computational Statistics
AS = Applied Statistics
Abraham Wald
R. Siegmund-Schultze (2003). Military Work in Mathematics 1914–1945: An Attempt at an International Perspective. Mathematics and War, edited by Bernhelm Booß-Bavnbek and Jens Høyrup, 23–82. DOI: 10.1007/978-3-0348-8093-0_2
2. Molecular biology: crash course
Does this matter?
- Genetics diseases, e.g. cancer
- Antibiotic resistance
- Virus outbreaks
- Food
- Ecology
- Information storage?
- . . .
Why is it suddenly a thing?

Why biological data science?
Because the skills are highly transferable:
- Nosy data
- Visualisation
- Communication
- High-performance computing
Formalising biology
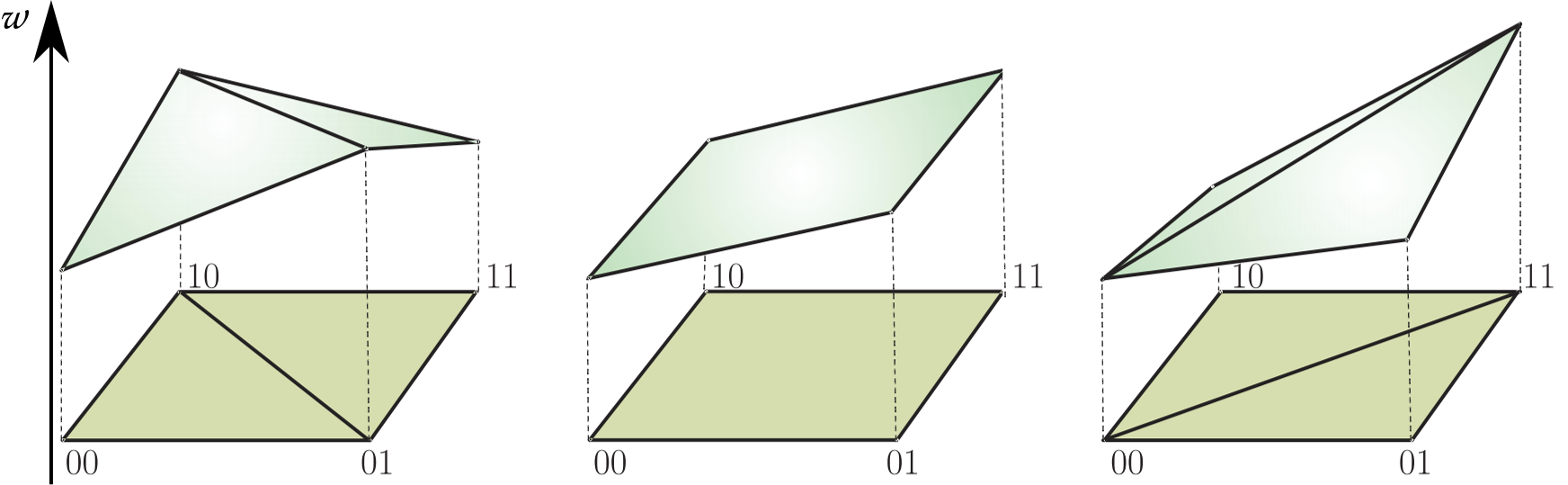
- Let $G$ be the set of binary (quaternary in reality) strings of length $n$.
- Elements of $G$ are called genotypes.
- A function $w: G \to \mathbb R^+$ is called a fitness landscape.
- For $g \in G$, $w(g)$ is called fitness of the genotype $g$.
Reality
Scalability: synthetic lethal pairs

How many pairs of genes are there in the human genome?
Gene-gene interactions

Epistasis
is defined as the deviation from the additive expectation of allelic effects: $$u_{11} = w_{00} + w_{11} - (w_{01} + w_{10})$$Understanding three-way interactions
Marginal epistasis?
$\small u_{\color{blue}{0}11} = w_{\color{blue}{0}00} + w_{\color{blue}{1}00} + w_{\color{blue}{0}11} + w_{\color{blue}{1}11} − (w_{\color{blue}{0}01} + w_{\color{blue}{1}01}) − (w_{\color{blue}{0}10} + w_{\color{blue}{1}10})$
Total three-way interaction?
$\small u_{111} = w_{000} + w_{011} + w_{101} + w_{110} - (w_{001} + w_{010} + w_{100} + w_{111})$
Conditional epistasis?
$\small e = w_{\color{blue}{0}00} − w_{\color{blue}{0}01} − w_{\color{blue}{0}10} + w_{\color{blue}{0}11}$
What if no (credible) fitness measurements are available?
Image: Wikipedia
Mutation fitness graph

Ogbunugafor et al. Malar. J. 2016
Rank orders. The simplest case.
$\small u_{11} = w_{00} + w_{11} - (w_{01} + w_{10})$
Exercise: Dyck word algorithm
$$ \begin{align} \small u_{011} =~ & w_{000} + w_{100} + w_{011} + w_{111} − \\ & w_{001} - w_{101} − w_{010} - w_{110} \end{align} $$
$$ w_{111} > w_{011} > w_{101} > w_{010} > w_{000} > w_{110} > w_{100} > w_{001} $$
$$ w_{111} > w_{011} > w_{100} > w_{000} > w_{001} > w_{101} > w_{010} > w_{110} $$
A way to quantify uncertainties!
Crona, Gavryushkin, Greene, and Beerenwinkel. Inferring genetic interactions from comparative fitness data. eLife, 2017.
Connection between rank orders and mutation graphs
$\small u_{11} = w_{00} + w_{11} - (w_{01} + w_{10})$
Theorem. A partial order (e.g. fitness graph) implies epistasis if and only if all linear extensions compatible with the partial order do.
Mutation graph

Mutation graph

Mutation graph

Mutation graph

Mutation graph

Lienkaemper, Lamberti, Drain, Beerenwinkel, and Gavryushkin. The geometry of partial fitness orders and an efficient method for detecting genetic interactions. Journal of Mathematical Biology, 2018.
4. Online algorithms to improve computational performance
- The traditional way is to make the algorithm more efficient
- When the same algorithm has to be re-run routinely, we can economise by making the algorithm slower and doing more!
- This approach is known as online
Online algorithms

Online algorithms

Online algorithms

Online algorithms

Online algorithms

Online algorithms

Online algorithms

Online algorithms

Online algorithms

Homework:
Find an efficient online algorithm to detect genetic interactions from
Acknowledgements

Thank you for your attention
and stay tuned
